Programming biological swarms
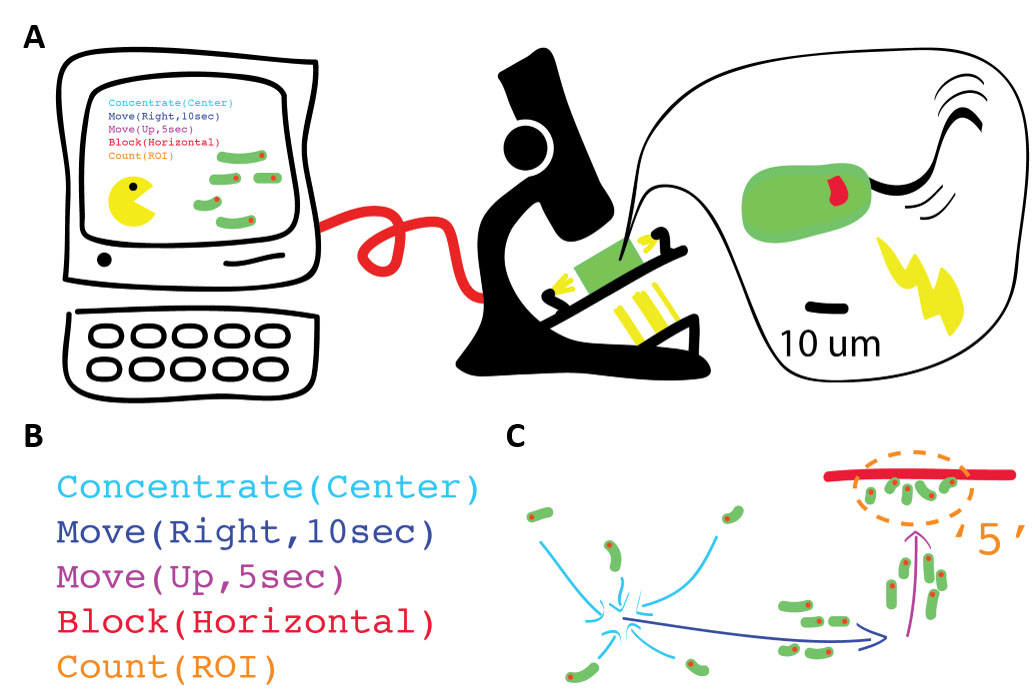
The prototype setup involves a microscope that is modified to enable light stimulation of the sample through a computer. This enables control over the swarm of Euglena gracilis (the sample), which in turn allows us to program the swarm.
The improvement in affordability, capabilities, and user interfaces of home computers led to a democratization of programming and software development to many people. A similar democratization of biology might be achieved if biological platforms for controlling and experimenting on living cells were available.
I built a system for controlling ensembles of the single-celled algae, Euglena gracilis. Euglena cells move in response to light stimuli (i.e. they are phototactic). In my system, local light stimuli can be applied through use of a standard pico projector with its signal demagnified, while global stimuli can be applied with LEDs outside of the field of view. Custom optics are used to enable imaging through a webcam, and custom software (in Processing language) is developed for easy control over the stimulus and cells.
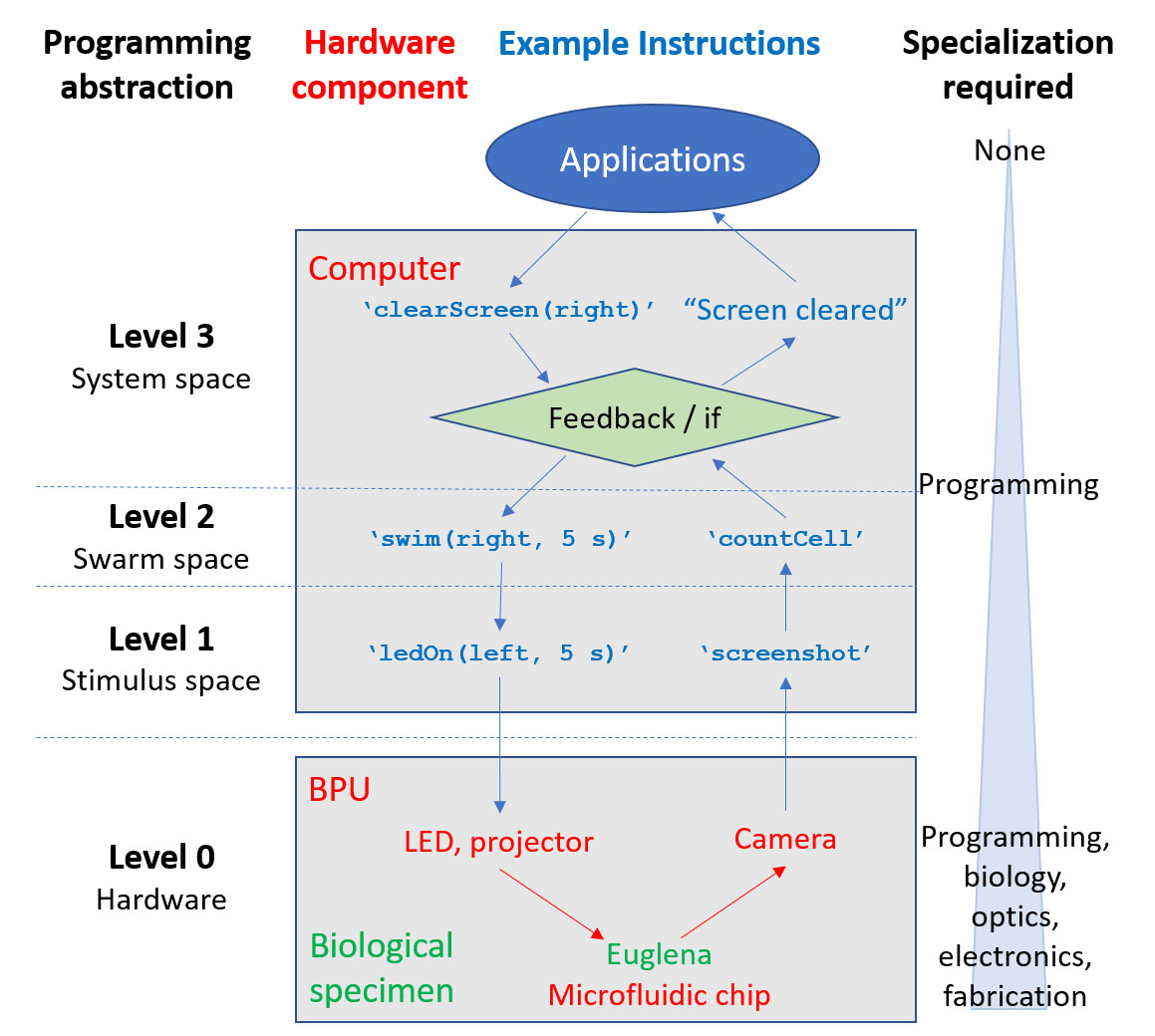
Programs for swarms can be at four different levels: hardware, stimulus level, swarm level, and system level.
Using this system, I explore the various levels of programming abstractions available to the user, finding that there are three natural levels to programming in biological computers. Ordered from the lowest to highest level of abstraction, they are the stimulus space, cell space, and system space. The upper two levels of abstraction depend upon the reliability of the cell responses as well as the ability for the computer to detect the state of the biological sample, which can be difficult to achieve due to the stochastic nature and large variability of biological specimen as well as limitations in computer vision. However, these upper layers of abstraction also enable easier programming as well as the development of broader programs which can be translated to other platforms (i.e. biological computers with different cells/stimulus combinations).
Additionally, I have used this system of hardware, software, and bioware to perform my own experiments on Euglena phototaxis and microswimmer swarm behaviors.
For further reading, see A. T. Lam, et al., Lab on a Chip (2017).